Such as the rise of immunecheckpoint inhibitors therapies, recent progresses for tumor therapy are the results of a more comprehensive analysis of the tumoral microenvironment (TME) especially the study of immune-tumor cell interactions.
Such discovery deeply modified our understanding of the TME and underlined the importance to integrate all the different cellular actors (tumor cells, immune cells, cancer-associated fibroblasts, endothelial cells etc.), their cellular state or sub cellular groups/compartments, their interactions and their localization within the TME.
For explorative purpose, it is essential to reach a significant number of individual biological parameter (cluster of differentiation, cytokine, transcription factors, etc.) to identify and characterize different cell subsets simultaneously.
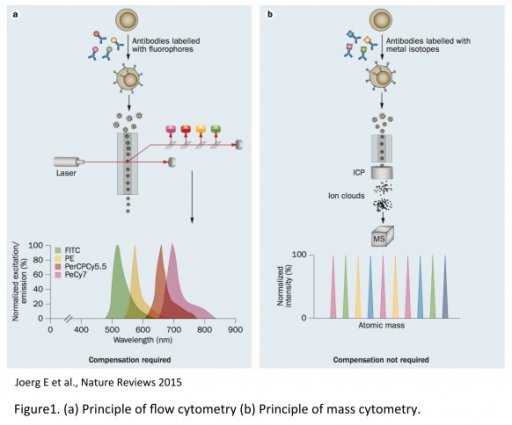
The CIM core provides the first Hyperion Imaging System (HIS). Developed by Fluidigm, the HIS is an imaging module associated to the Helios, a 3rd generation of CyTOF, which allows single cell suspension (SCS) analysis and tissue imaging using the mass spectrometry technology. Mass cytometry uses metal-labeled primary antibodies detected by mass spectrometry (Time Of Flight, TOF). This method overcomes then many of fluorescent-based technology limitations such as spectral overlap, auto fluorescence, dye instability or photobleaching. Such limitations make more difficult to distinguish multiple optical signals especially as more parameters are measured in a single experiment.
In contrast, elemental mass spectrometry is able to discriminate isotopes of different atomic weights with high accuracy. This fundamental difference enables significantly more cellular features to be assayed simultaneously using a mass-based platform
(Reviewed in Spitzer et al., Cell 2016).
Mass cytometry supports then high multiplexing with up to 50 stable isotopes currently available for 135 opened channels. Isotopes can be conjugated to antibodies or probes for protein levels, posttranslational modifications, proteolysis products, cell signaling, mRNA transcripts or DNA synthesis quantification from a single experiment.
Data generated can then be analyzed and interpreted the same way as FACs analysis (FCS files).
The Hyperion allows similar multiparametric staining on tissue section. An UV laser ablates the tissue section (1µm2) and events are then analyzed by the Helios.
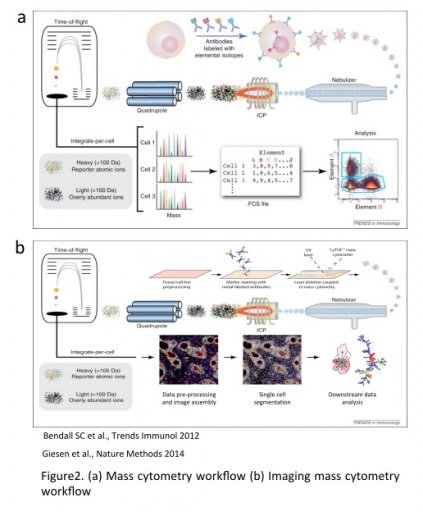
The combination of the SCS analysis together with the tissue imaging allows three level of information :
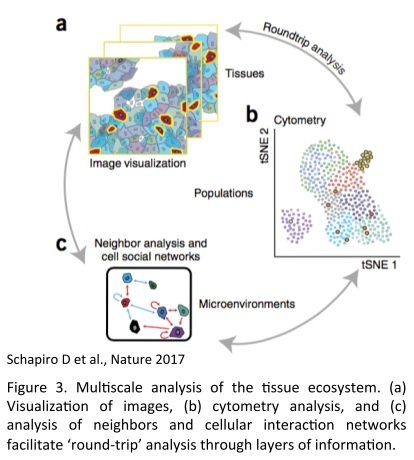
- The identification of cell types, biological processes (functionality, proliferation, apoptosis) and their particular distribution report on the spatial composition of the TME
- Cell segmentation and single cell unsupervised-analysis (Principal Component Analysis, span tree, clustering) allow the discovery of unexpected cell subtypes and heterogeneity in an unbiased manner. Subcellular compartment and biological activities can be assigned based on the presence or absence of markers
- The spatial view and cell segmentation allow to put into their environmental context each cell type and its functional state. Those states can be related to cell types, states and neighborhoods and eventually reconstitute the cellular social network of populations of interest.
The applications of mass-based SCS analysis and tissue imaging are numerous and varied. Beyond a deep and comprehensive study of the TME, biomarker and drug discovery, pharmacokinetics of drugs into the tumor, therapeutic assessment of drug combination are also feasible. Furthermore, data generated can be compared against or integrated to data generated with ‘omics’ data such as genotype, transcriptome, proteome, matabolome or imaging.